last update : january 2015
Public data retrieved in 2014
AT_CHLORO
A database dedicated to the subplastidial localization of proteins
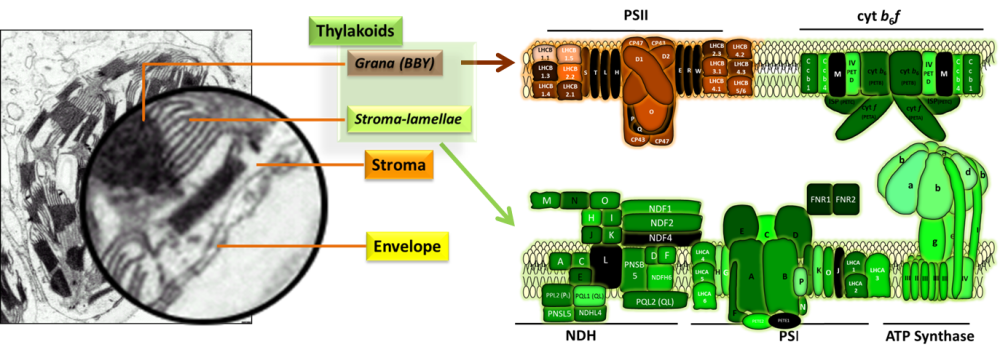
The AT_CHLORO database stores sub-plastidial (envelope, stroma, thylakoids; Ferro et al., Mol Cell Proteomics 2010; Bruley et al., Frontiers Plant Sci 2012) and sub-thylakoidal (Grana, stroma-lamellae) localization of The AT_CHLORO database links to other public web sites (TAIR, PPDB, AtProteome, SUBA, POGs, Aramemnon)
The AT_CHLORO database stores information for proteins that have been identified in different sub-fractions obtained from
A. thaliana chloroplasts (Ferro et al., 2010).
Users can (i) consult the whole database, (ii) retrieve proteins that are localized in a sub-plastidial compartment (iii) or choose a set of proteins with their AGI accession numbers (Searching option).
If you find the AT_CHLORO database useful, please cite one of the following publications:
The MASCP Gator (Joshi et al., Plant Physiol. 2011), the aggregation portal for the visualization of Arabidopsis proteomics data, gathers information from a variety of online proteomics resources including AT_CHLORO.
How to use the AT_CHLORO database
A description of the features can be found in the help page.
Citation
In addition AT_CHLORO also stores plastidial calmodulin-binding proteins (Dell’Aglio et al., 2013)
Methods used for subcellular and subplastidial fractionation of Arabidopsis chloroplast are described in the following publications:
- Salvi D, Moyet L, Seigneurin-Berny D, Ferro M, Joyard J, Rolland N. Preparation of envelope membrane fractions from Arabidopsis chloroplasts for proteomic analysis and other studies. Methods Mol Biol. 2011;775:189-206.
- Salvi D, Rolland N, Joyard J, Ferro M. Purification and proteomic analysis of chloroplasts and their sub-organellar compartments. Methods Mol Biol. 2008;432:19-36. doi: 10.1007/978-1-59745-028-7_2.
The AT_CHLORO database was used to explore the subplastidial localization of specific metabolic pathways
- Joyard J, Ferro M, Masselon C, Seigneurin-Berny D, Salvi D, Garin J, Rolland N. Chloroplast proteomics and the compartmentation of plastidial isoprenoid biosynthetic pathways. Mol Plant. 2009 Nov;2(6):1154-80.
- Joyard J, Ferro M, Masselon C, Seigneurin-Berny D, Salvi D, Garin J, Rolland N. Chloroplast proteomics highlights the subcellular compartmentation of lipid metabolism. Prog Lipid Res. 2010 Apr;49(2):128-58.
- Rolland N, Curien G, Finazzi G, Kuntz M, Maréchal E, Matringe M, Ravanel S, Seigneurin-Berny D. The biosynthetic capacities of the plastids and integration between cytoplasmic and chloroplast processes. Annu Rev Genet. 2012;46:233-64.
EDyP laboratory
PCV laboratory
Web Master : veronique.dupierris@cea.fr (EDyP); sylvain.bournais@cea.fr (EDyP)
Contacts: norbert.rolland@cea.fr (PCV); myriam.ferro@cea.fr (EDyP)